v18.01
GTRD
Gene Transcription Regulation Database
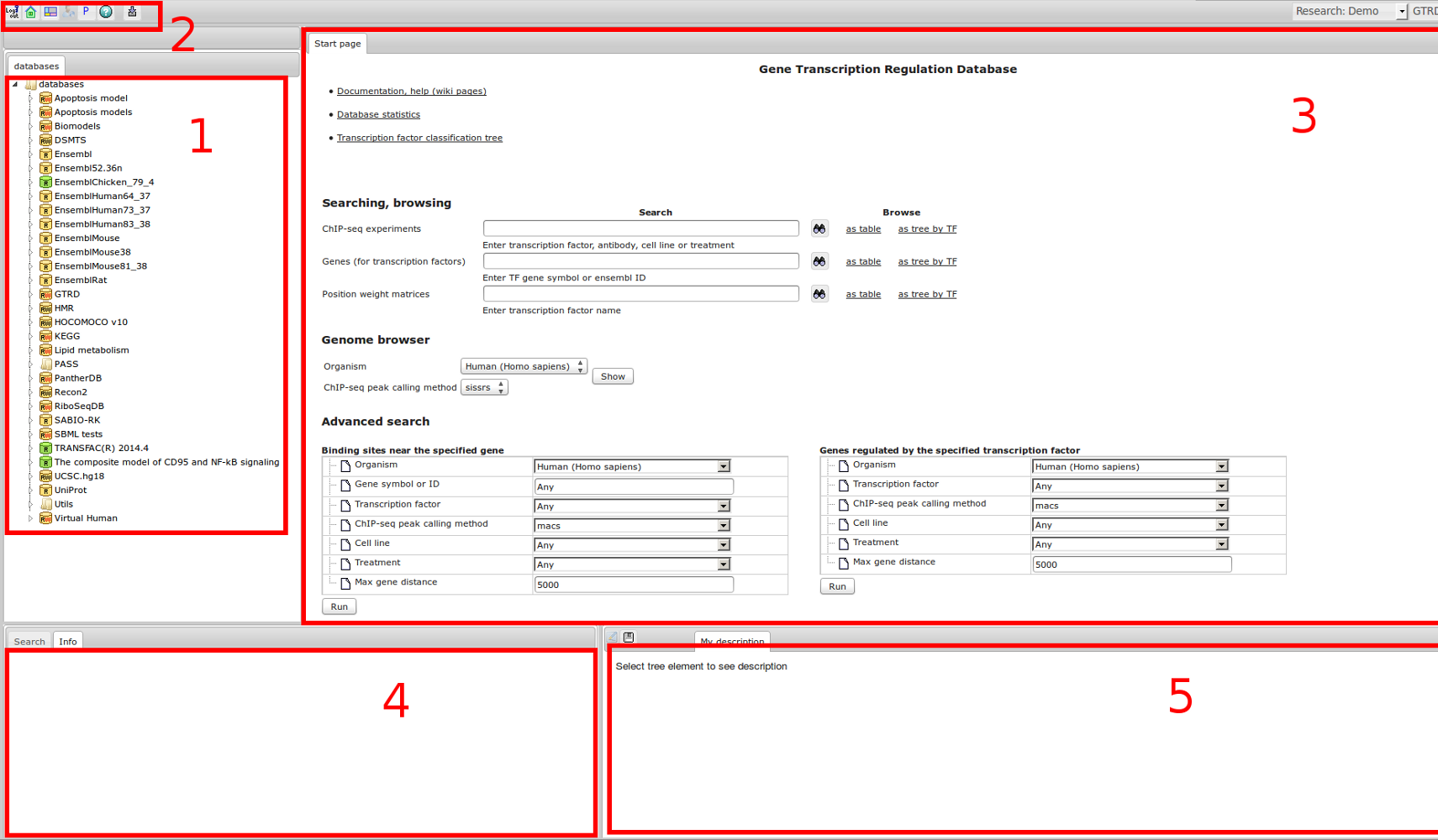
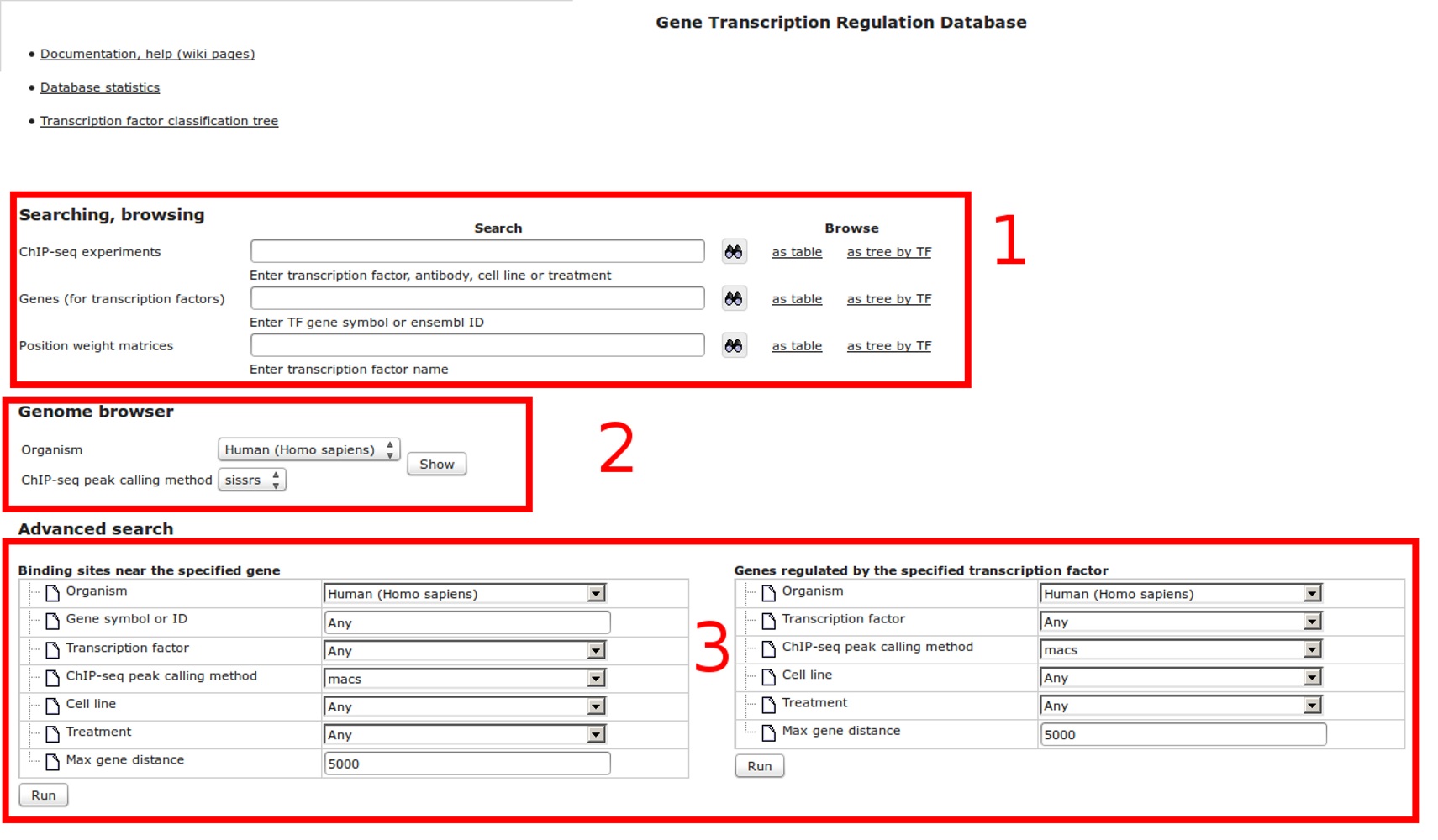
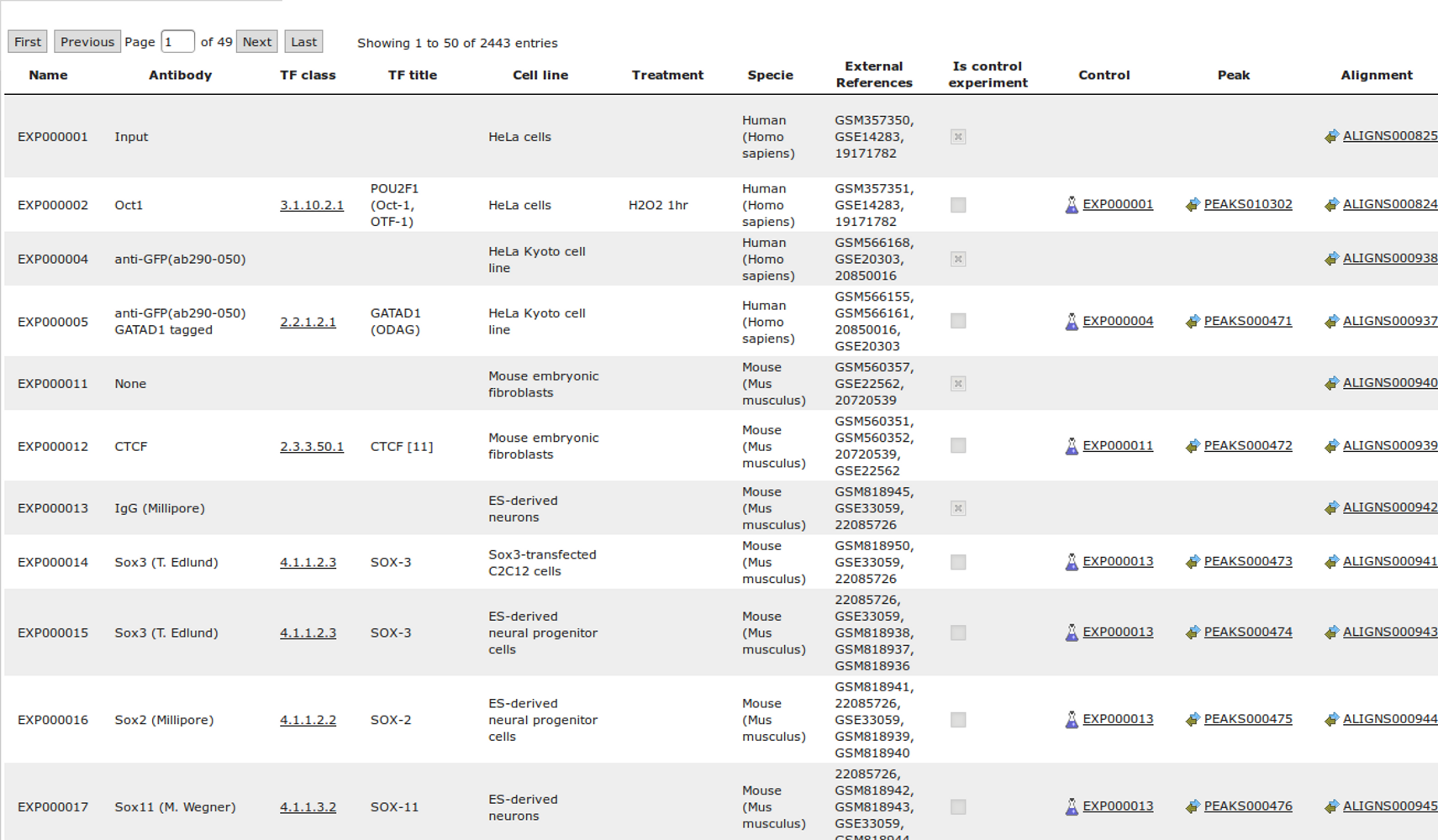
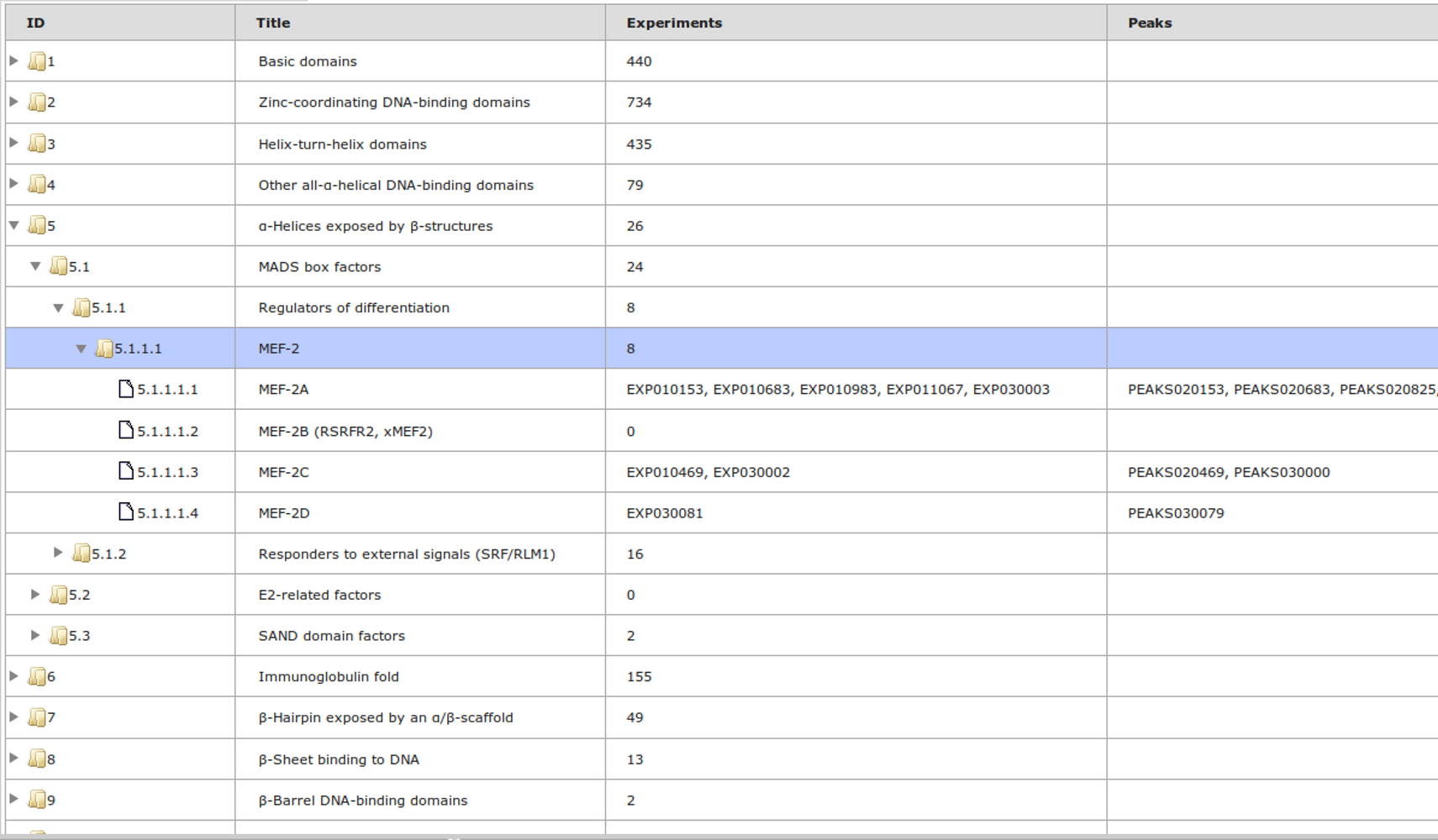
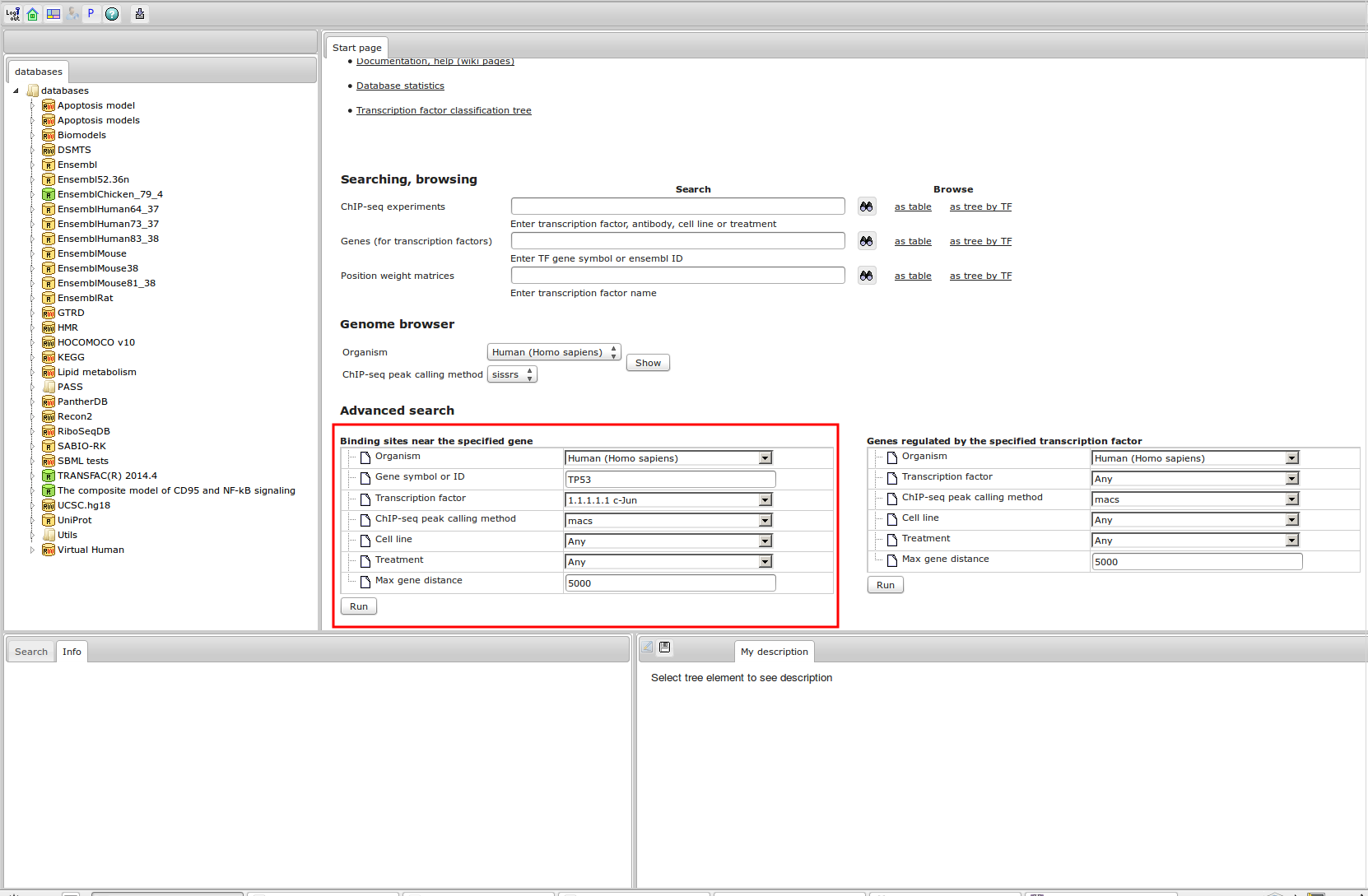
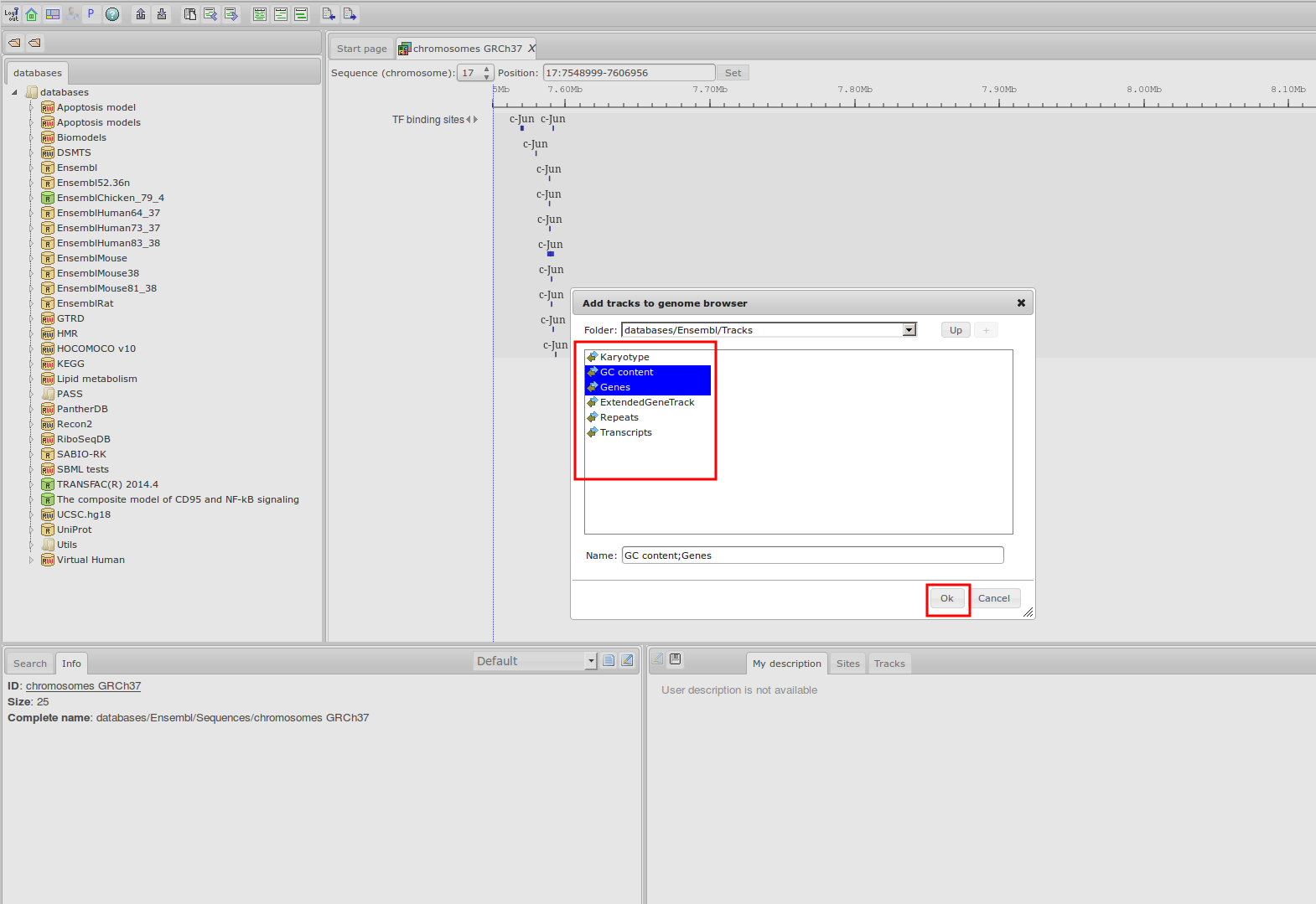
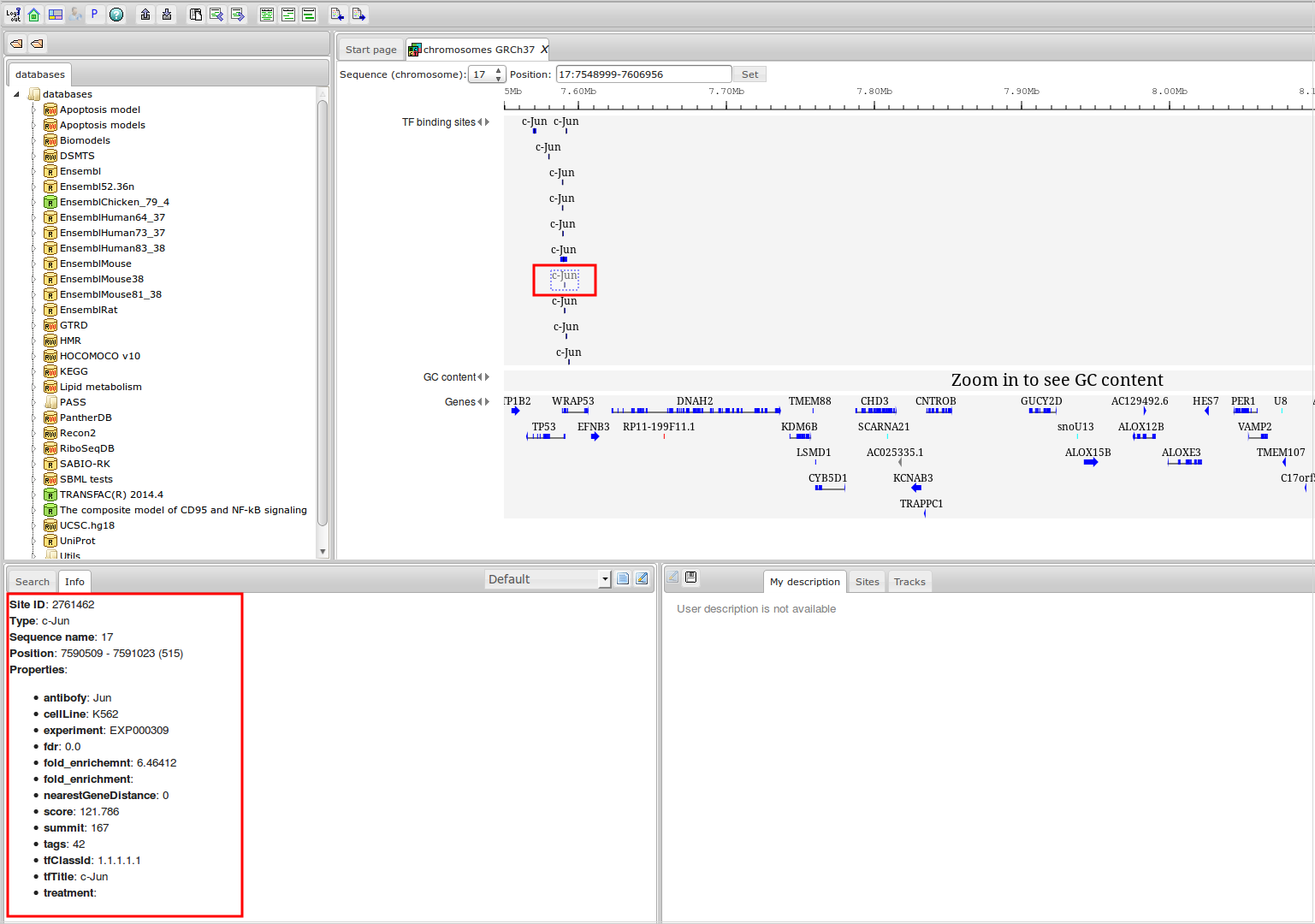
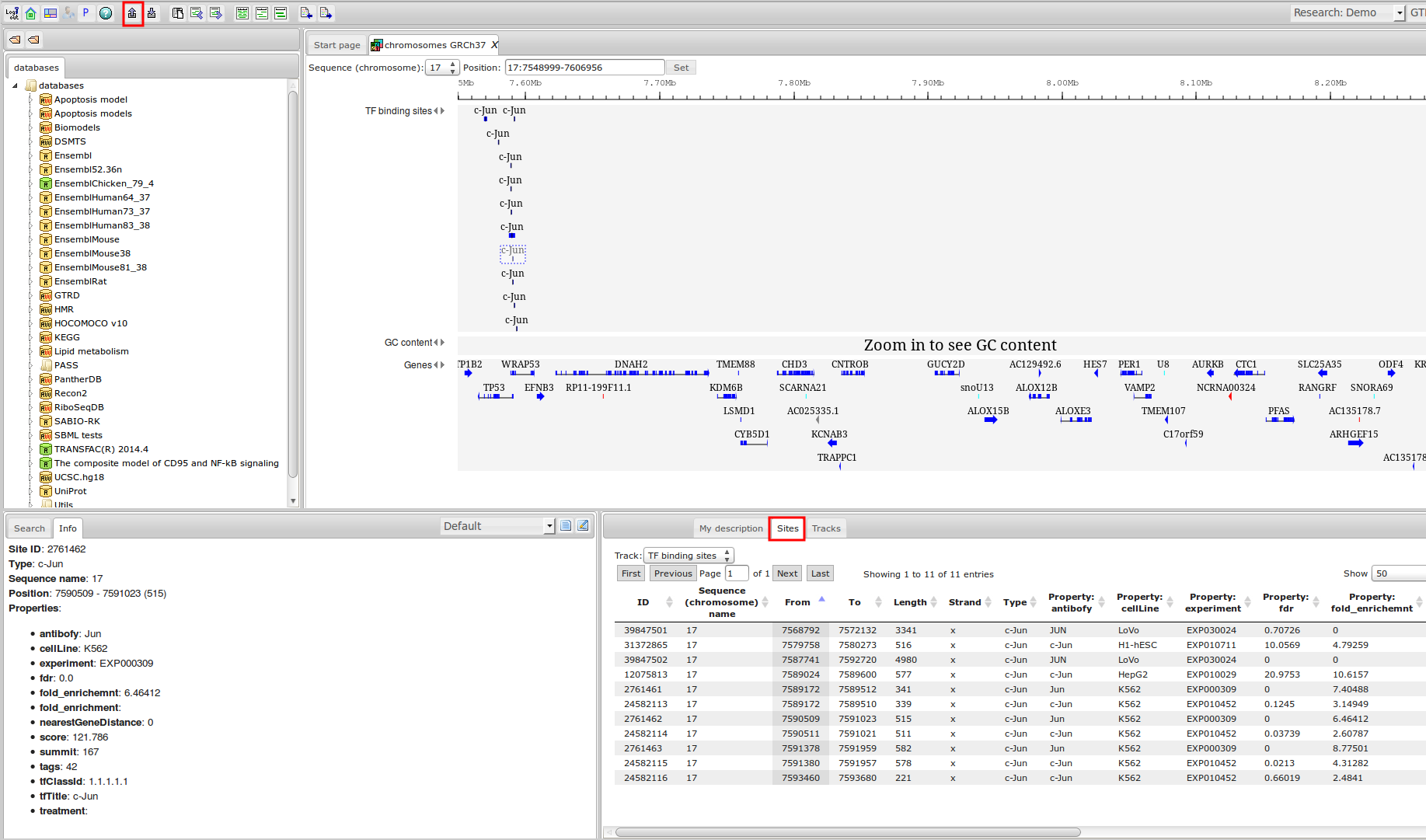
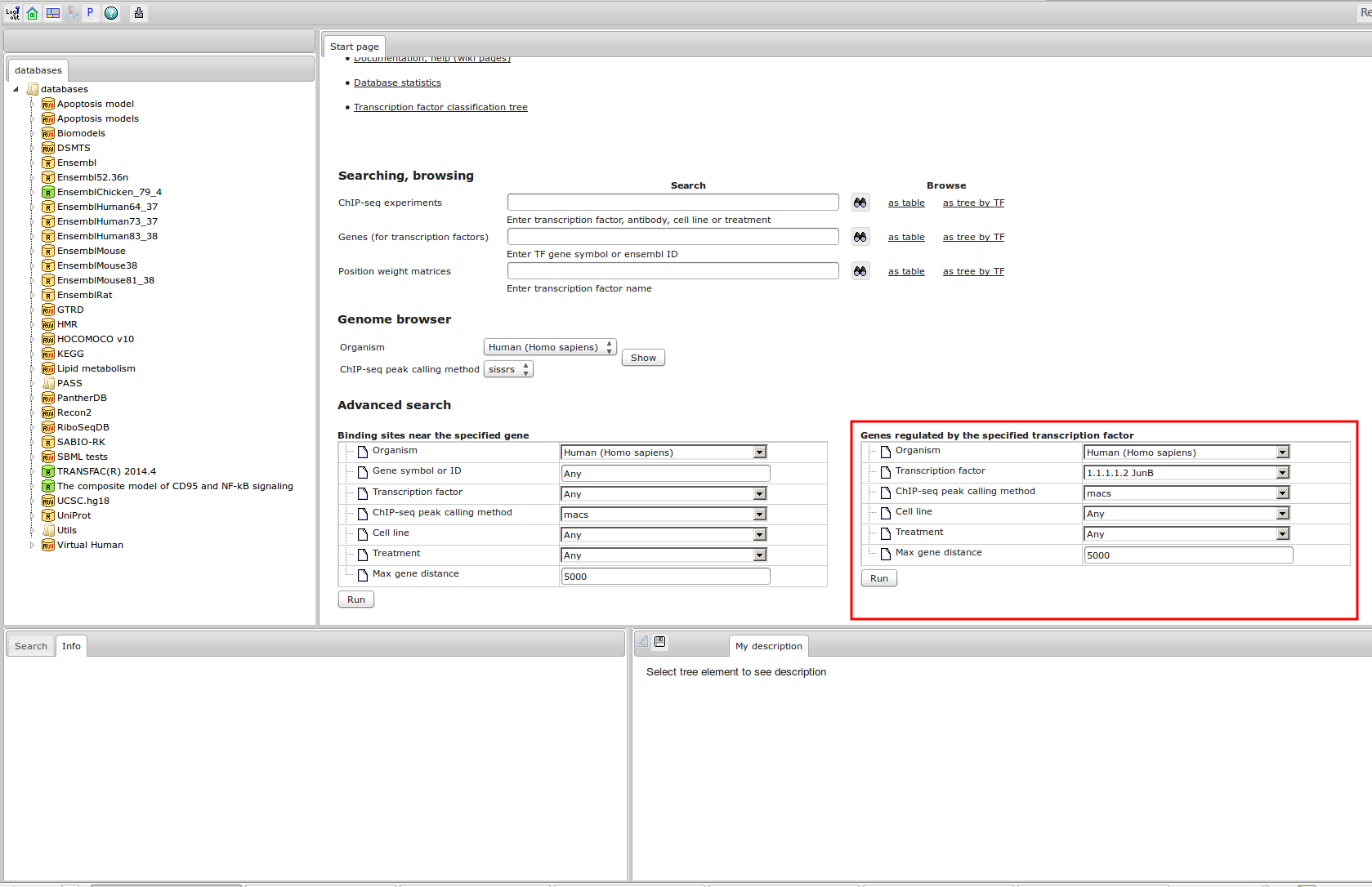
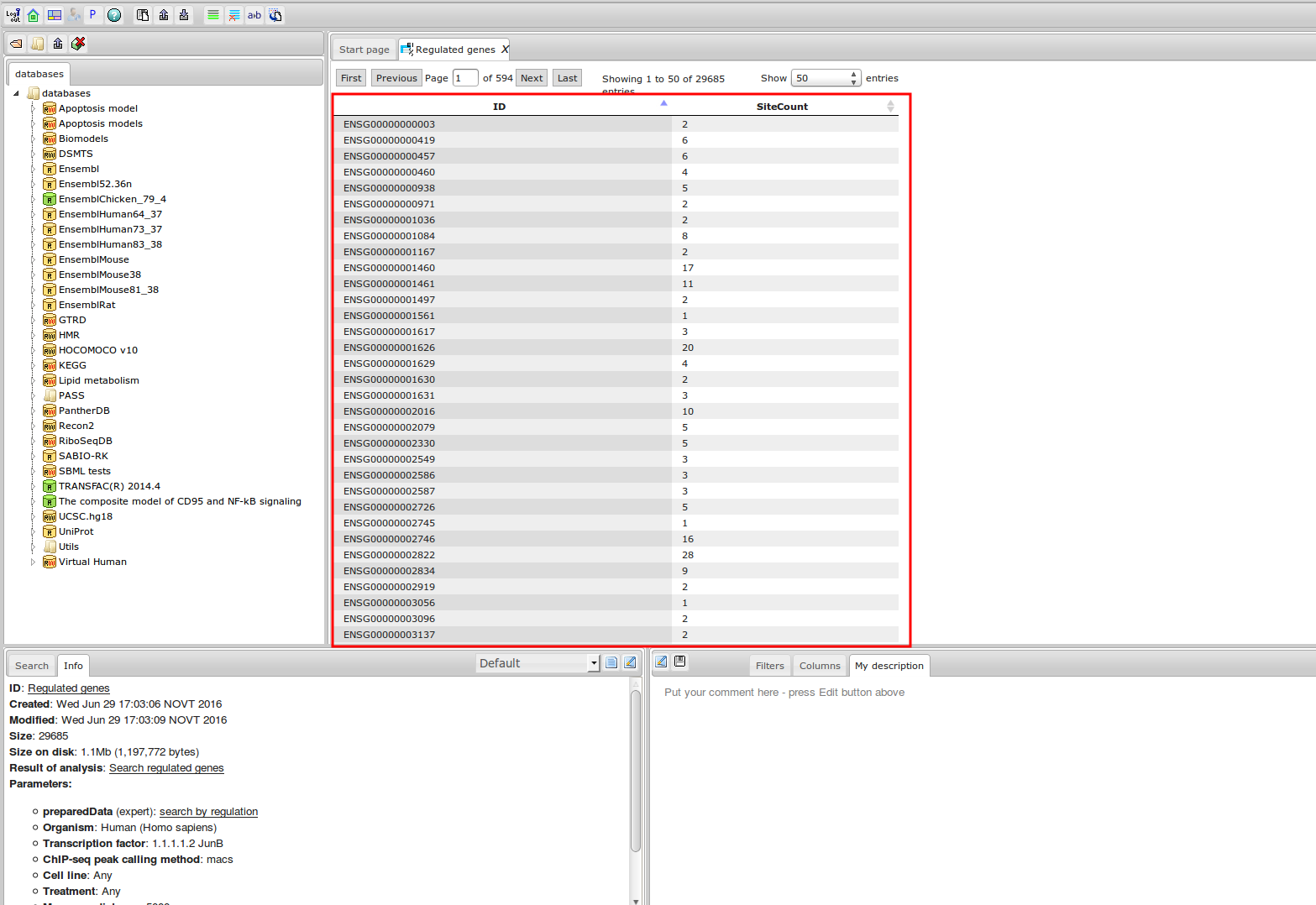
The most complete collection of uniformly processed ChIP-seq data to identify transcription factor binding sites for human and mouse. Convenient web interface with advanced search, browsing and genome browser based on the BioUML platform. For support or any questions contact ivan@dote.ru
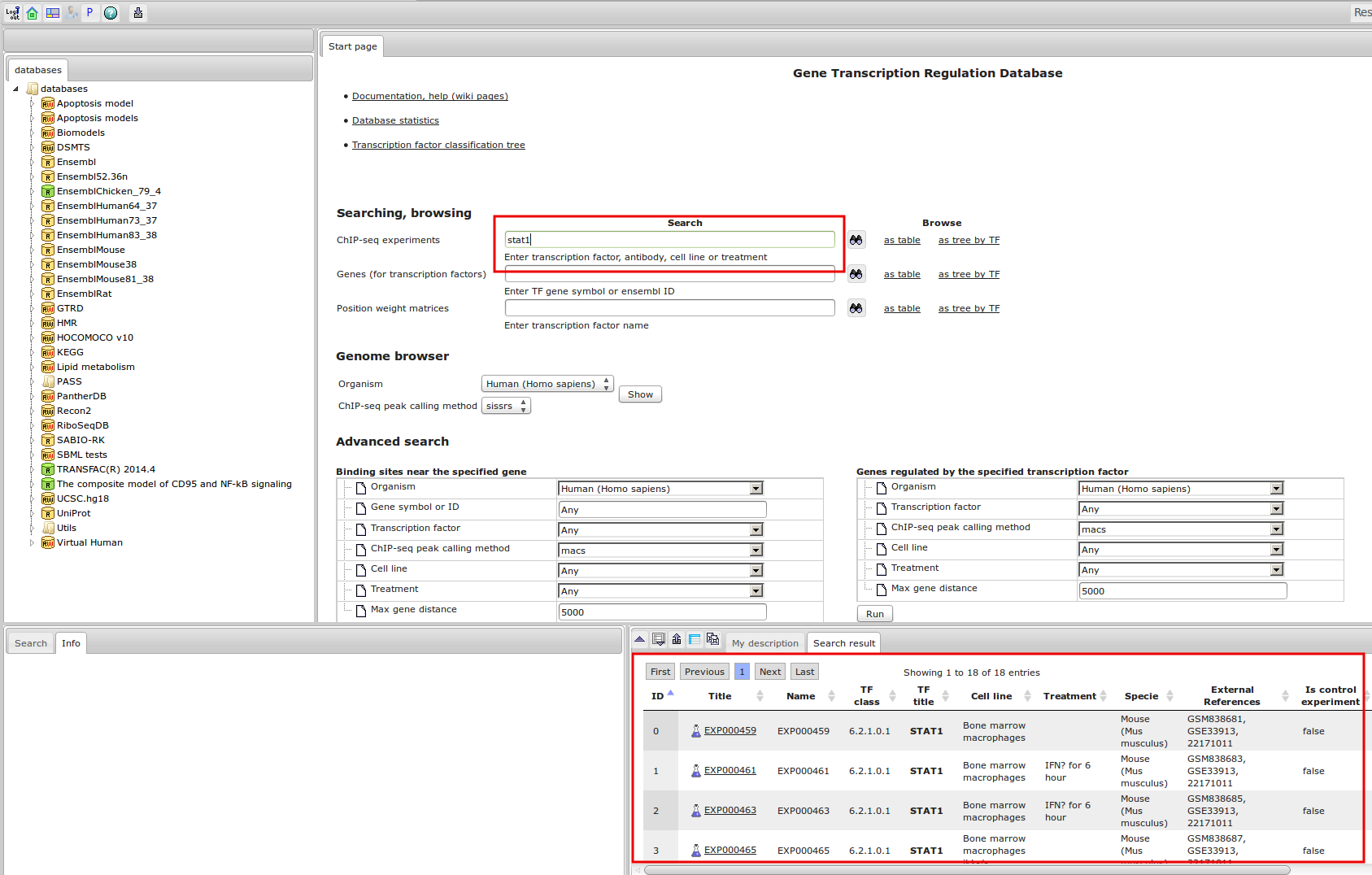
Workflow
How was it constructed?

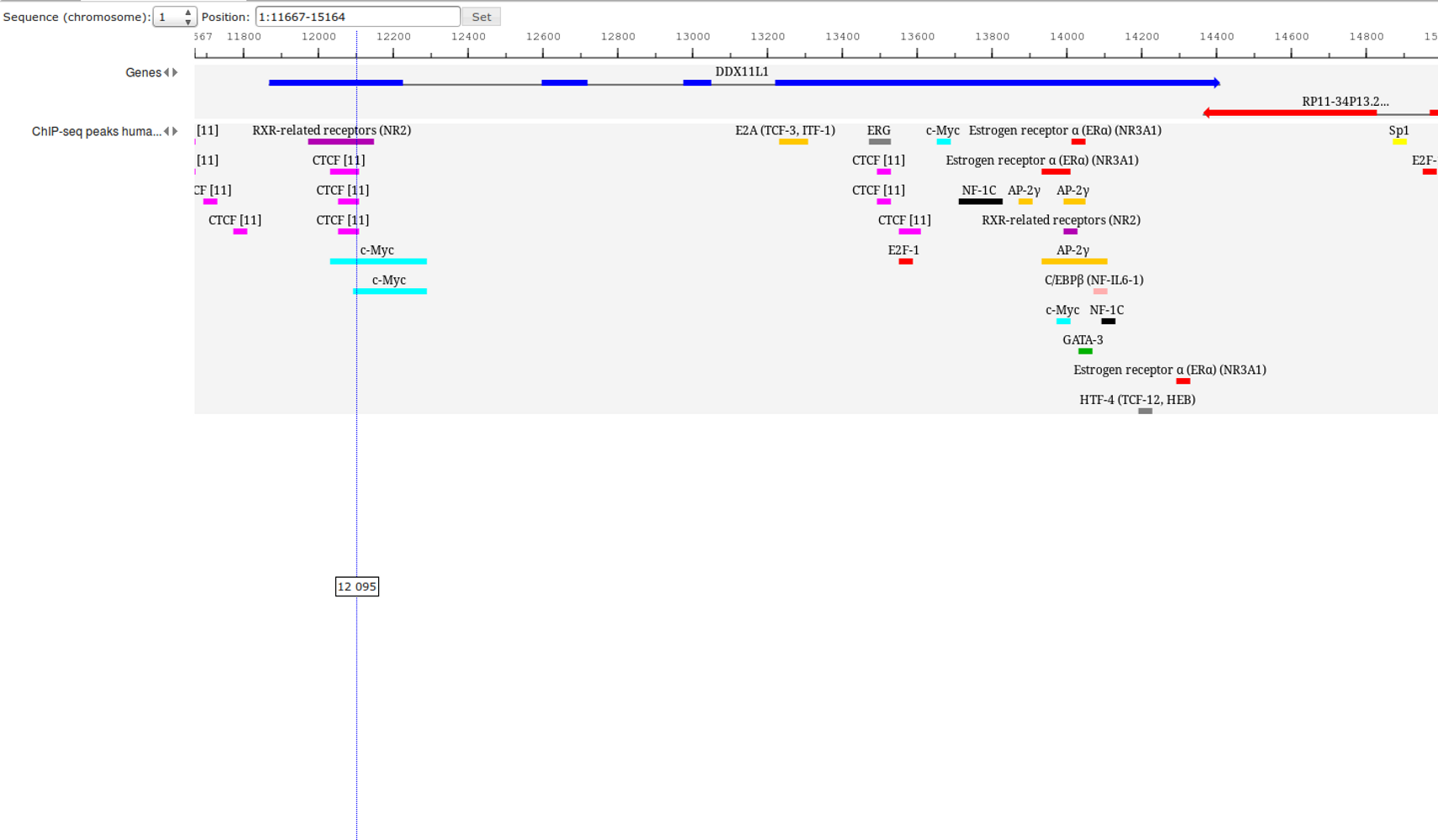
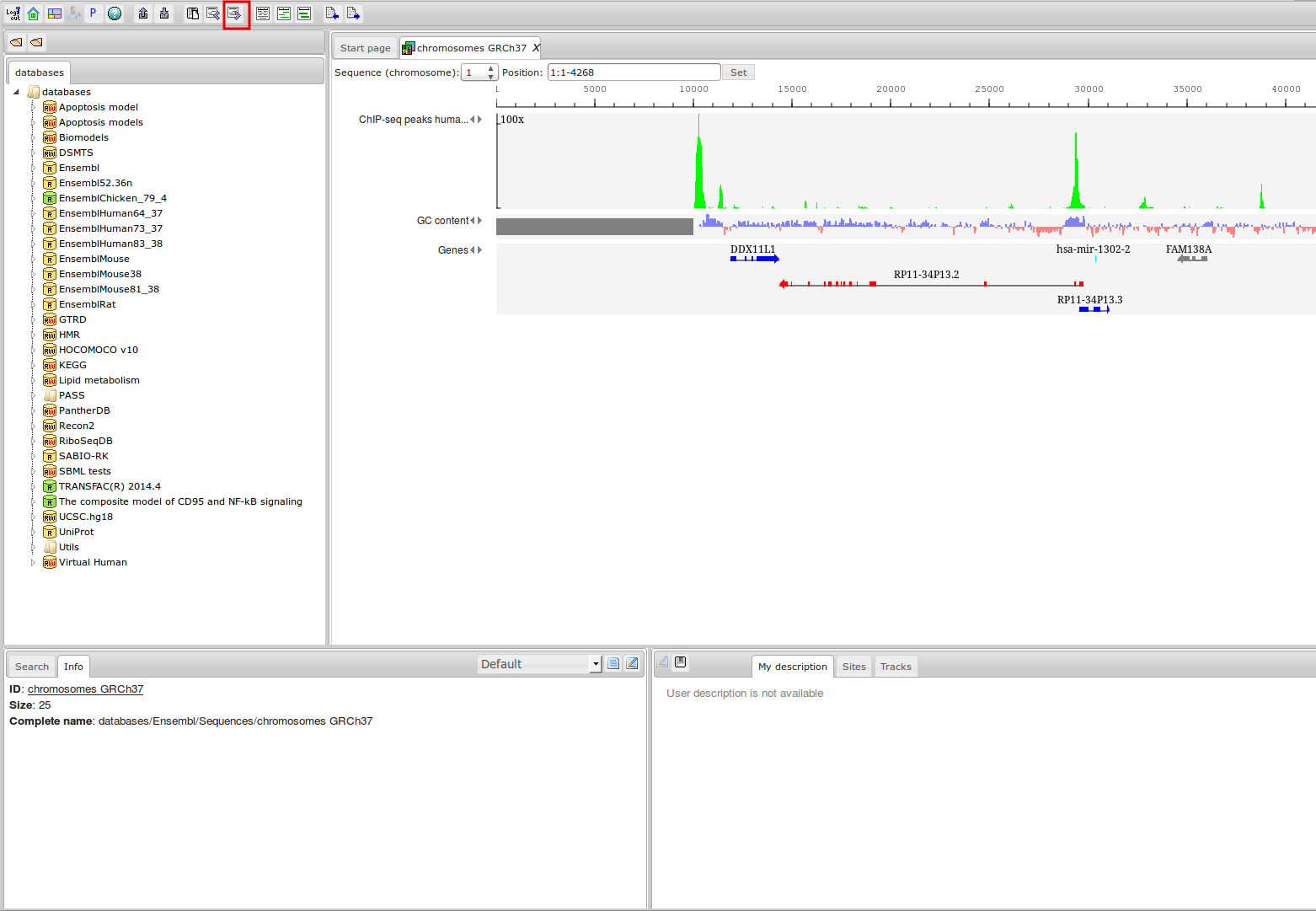
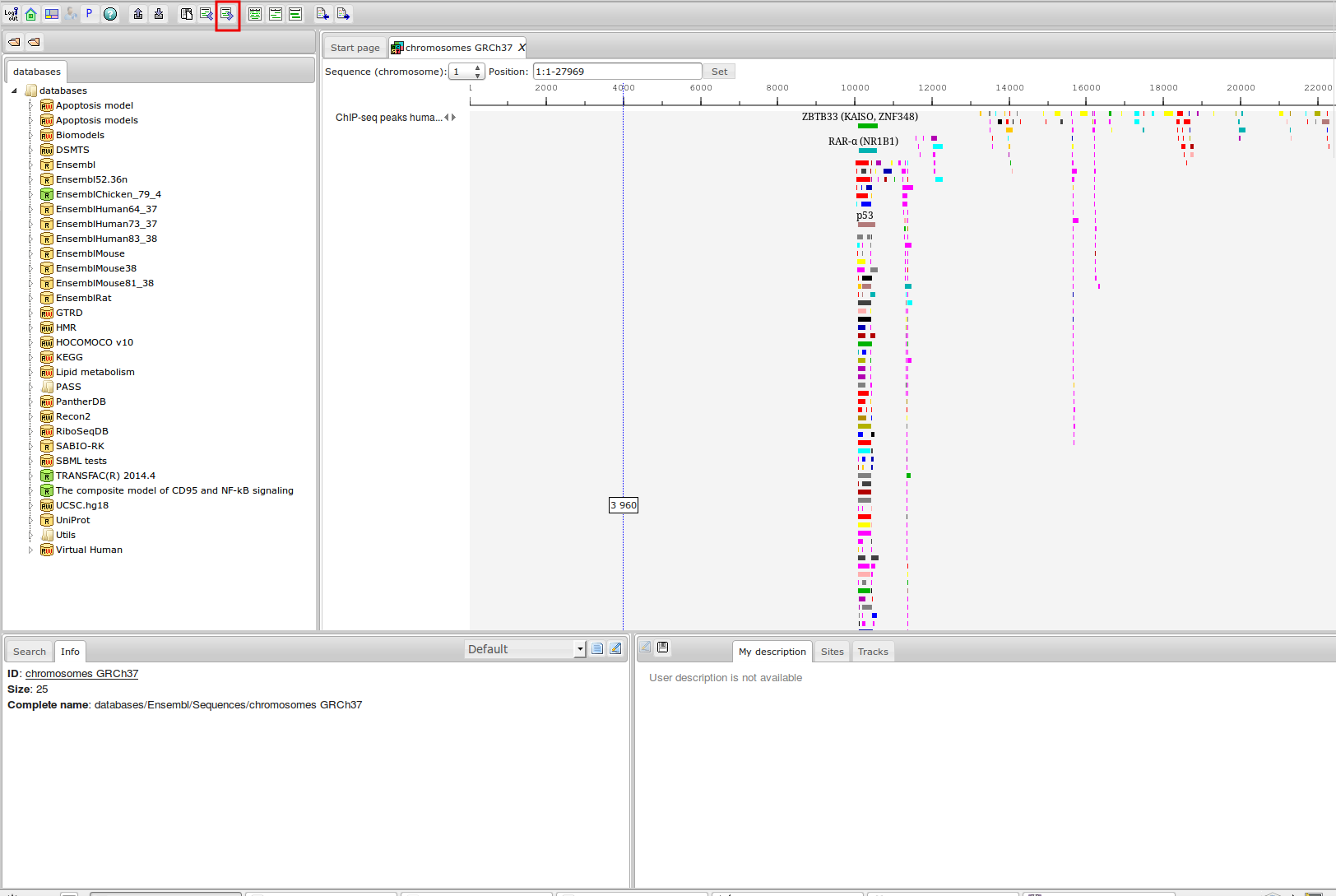
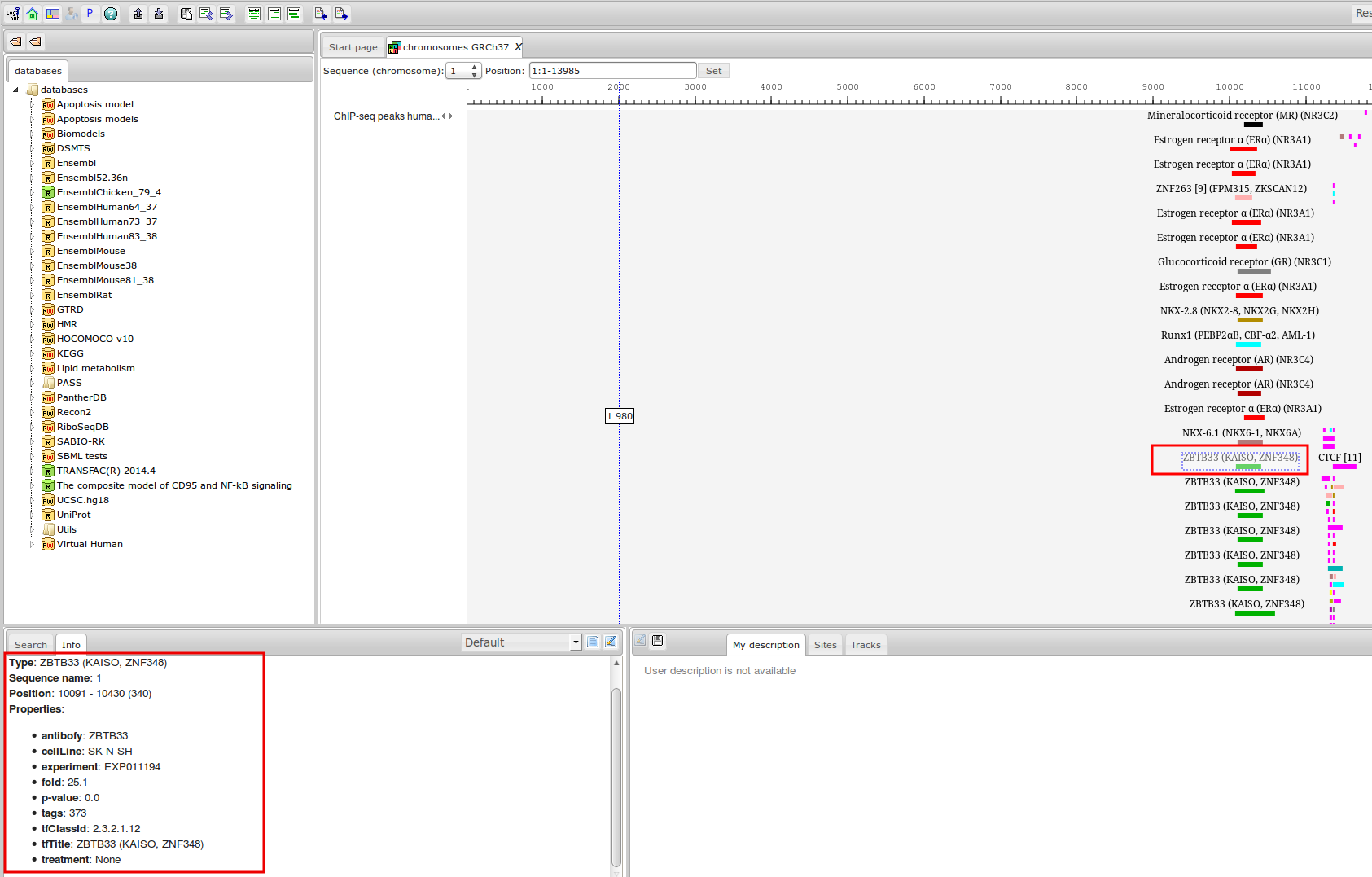
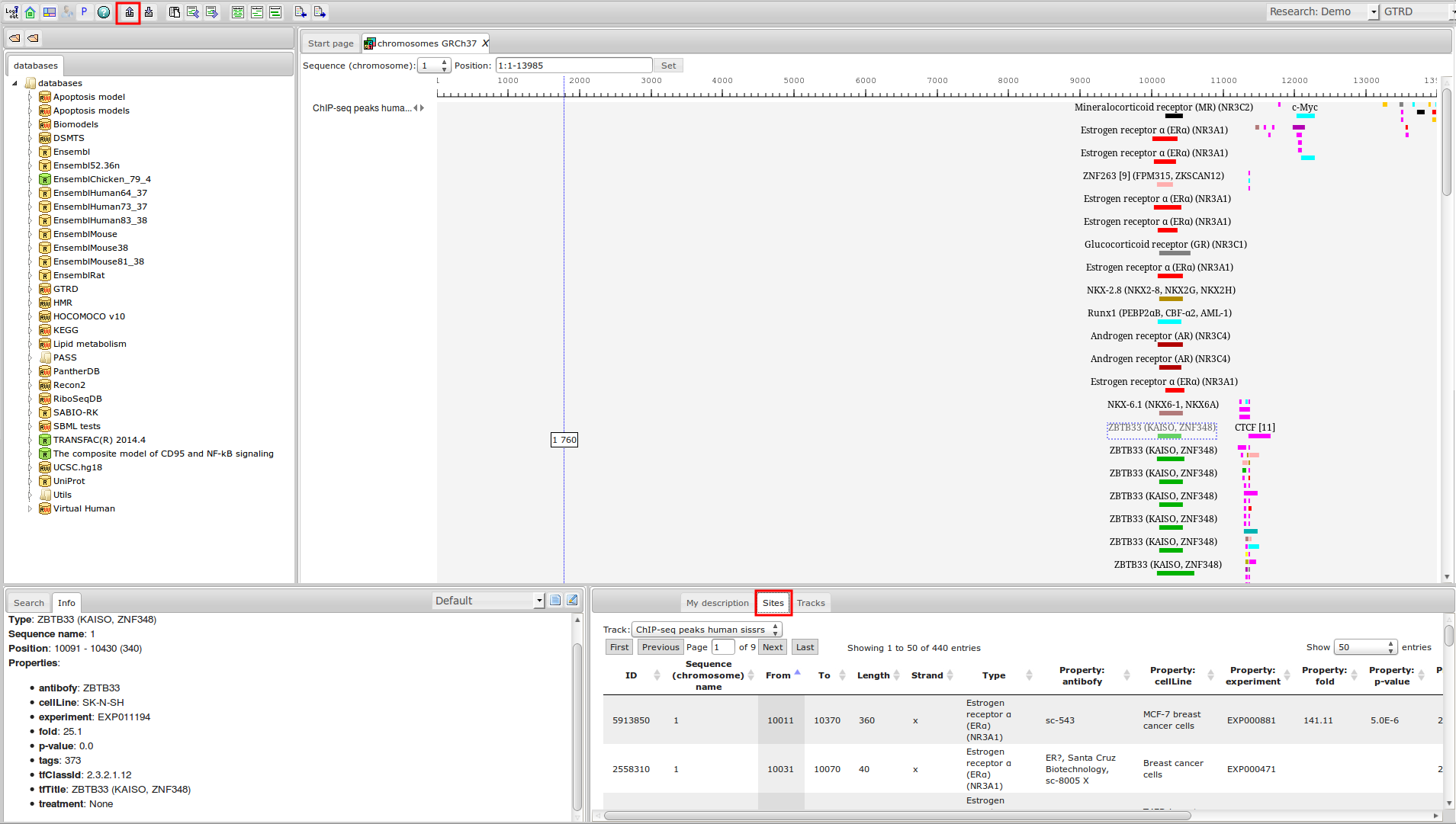
ChIP-seq experiment information and raw data were collected from publically available sources. Sequenced reads were aligned using Bowtie2 and ChIP-seq peaks were called using 4 different methods. Peaks were merged into clusters and metaclusters to produce non-redundant set of transcription factor binding sites.
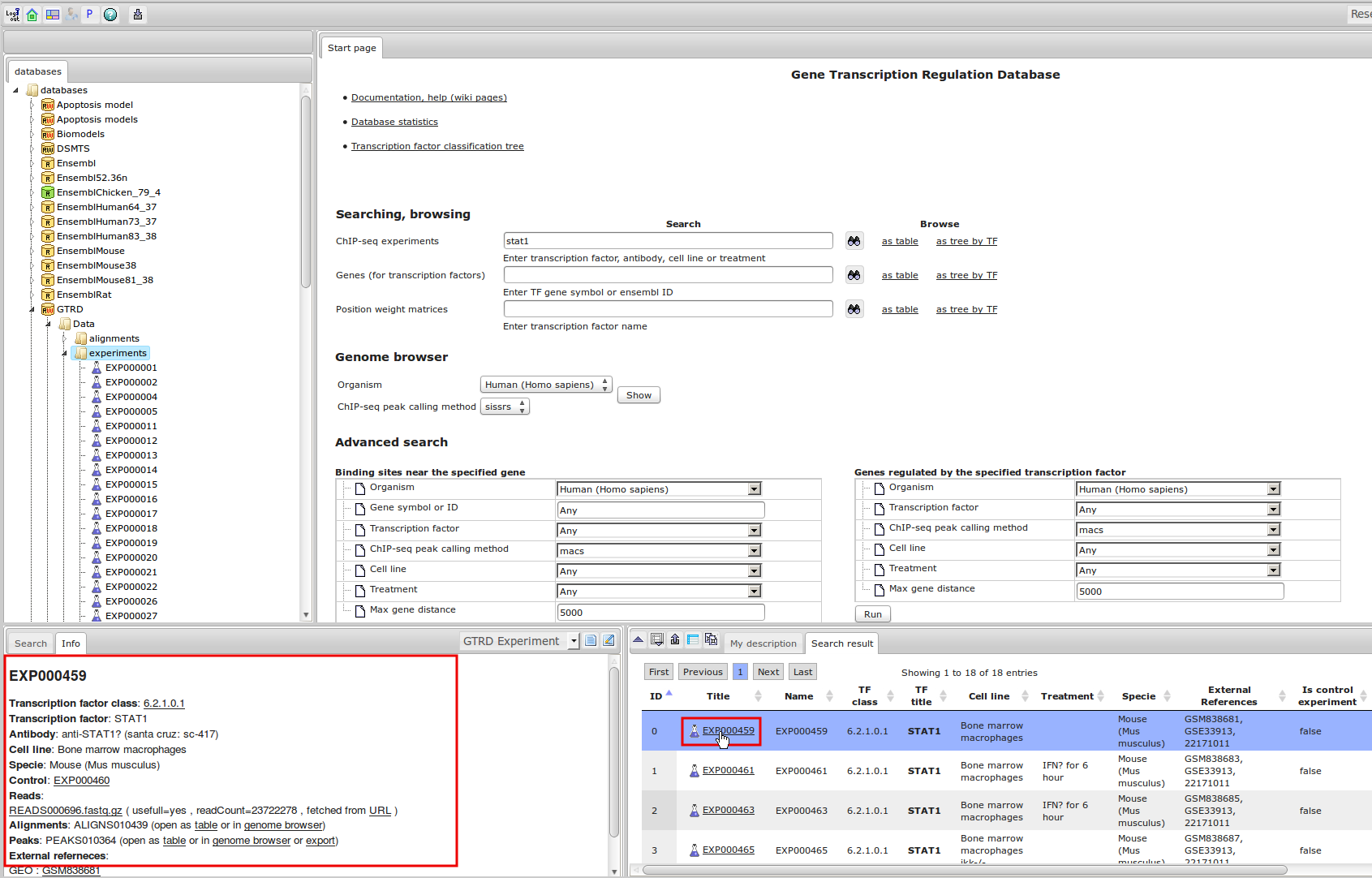
Learn more »Statistics
version 18.01
- 3340 new 12168 ChIP-seq experiments
- 53 new 766 Transcription factors
- 27% new 508 023 543 763 ChIP-seq reads
- 29% new 335 707 127 761 Reads aligned
- 31% new 961 923 622 ChIP-seq peaks
- 20% new 527 498 674 Clusters
- 14% new 70 338 813 Metaclusters
How to cite
License
Users may freely use the GTRD database for non-commercial purposes as long as they properly cite it. If you intend to use GTRD for a commercial purpose, please contact ivan@dote.ru to arrange a license.
Studies citing GTRD
- Genome-Wide Scanning of Gene Expression. Reference Module in Life Sciences. 2018
- Multi-omic Directed Networks Describe Features of Gene Regulation in Aged Brains and Expand the Set of Genes Driving Cognitive Decline Front. Genet., 09 August 2018
- Analysis of diet-induced differential methylation, expression, and interactions of lncRNA and protein-coding genes in mouse liver. Sci Rep. 2018 Aug 1;8(1):11537.
- Systems analysis of dilated cardiomyopathy in the next generation sequencing era. Wiley Interdiscip Rev Syst Biol Med. 2018 Jul
- Hypoxia and serum deprivation induces glycan alterations in triple negative breast cancer cells. Biol Chem. 2018 Jun 27
- Hormone-responsive genes in the SHH and WNT/β-catenin signaling pathways influence urethral closure and phallus growth. Biol Reprod. 2018 May 14.
- The BaMM web server for de-novo motif discovery and regulatory sequence analysis. Nucleic Acids Research, 28 May 2018
- Transcription Factor Databases. Reference Module in Life Sciences. 2018
- Core signaling pathways in ovarian cancer stem cell revealed by integrative analysis of multi-marker genomics data. PLoS One. 2018 May 3.
- Computational master-regulator search reveals mTOR and PI3K pathways responsible for low sensitivity of NCI-H292 and A427 lung cancer cell lines to cytotoxic action of p53 activator Nutlin-3. BMC Medical Genomics, 13 February 2018.
- MnSOD/SOD2 in Cancer: The Story of a Double Agent. Reactive Oxygen Species, Feb 2018.
- The Role of Indoleamine-2,3-Dioxygenase in Cancer Development, Diagnostics, and Therapy. Front. Immunol., 31 January 2018
-
Eukaryotic and prokaryotic promoter databases as valuable tools in exploring the regulation of gene transcription: a comprehensive overview.
Gene. 2017 Nov 2. -
HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis.
Nucleic Acids Res. 2017 Nov 11. - ReMap 2018: an updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-seq experiments Nucleic Acids Res. 2017 Nov 8.
- DMS-Seq for In Vivo Genome-wide Mapping of Protein-DNA Interactions and Nucleosome Centers. Cell Reports 2017 Oct 3;21(1):289-300.
- EpiDenovo: a platform for linking regulatory de novo mutations to developmental epigenetics and diseases. Nucleic Acids Research 2017, gkx918
- Genetic variants in ADAMTS13 as well as smoking are major determinants of plasma ADAMTS13 levels. Blood Advances 2017 1:1037-1046;
- Master-regulators driving resistance of non-small cell lung cancer cells to p53 reactivator Nutlin-3. Virtual Biology 2017, 0(4), 1-31.
- Discovering relationships between nuclear receptor signaling pathways, genes, and tissues in Transcriptomine. Sci Signal. 2017 Apr 25;10(476).
- A comprehensive review of web-based non-coding RNA resources for cancer research. Cancer Lett. 2017 Aug 18;407:1-8.
- RUNX1 promote invasiveness in pancreatic ductal adenocarcinoma through regulating miR-93. Oncotarget 2017 Aug 24